Build your own mass spectrometry analysis pipeline in Python using matchms — part I | by Florian Huber | Netherlands eScience Center
tanimoto_similarities.py: A Python script to calculate Tanimoto similarities of multiple compounds using RDKit. — Bioinformatics Review

Pyteomics 4.0: Five Years of Development of a Python Proteomics Framework | Journal of Proteome Research
Python: Find the type of the progression and the next successive member of a given three successive members of a sequence - w3resource
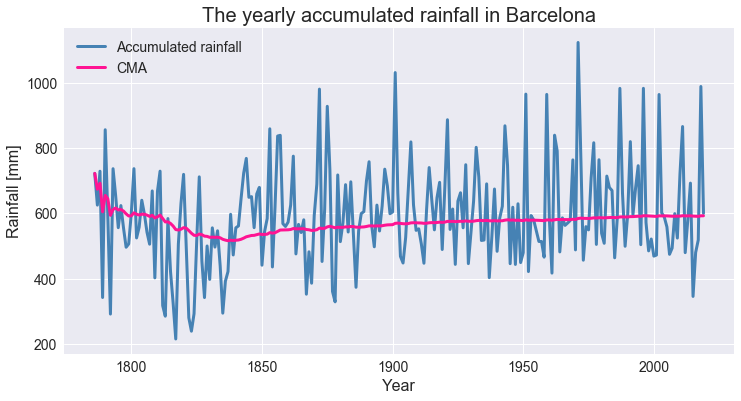
Moving averages with Python. Simple, cumulative, and exponential… | by Amanda Iglesias Moreno | Towards Data Science
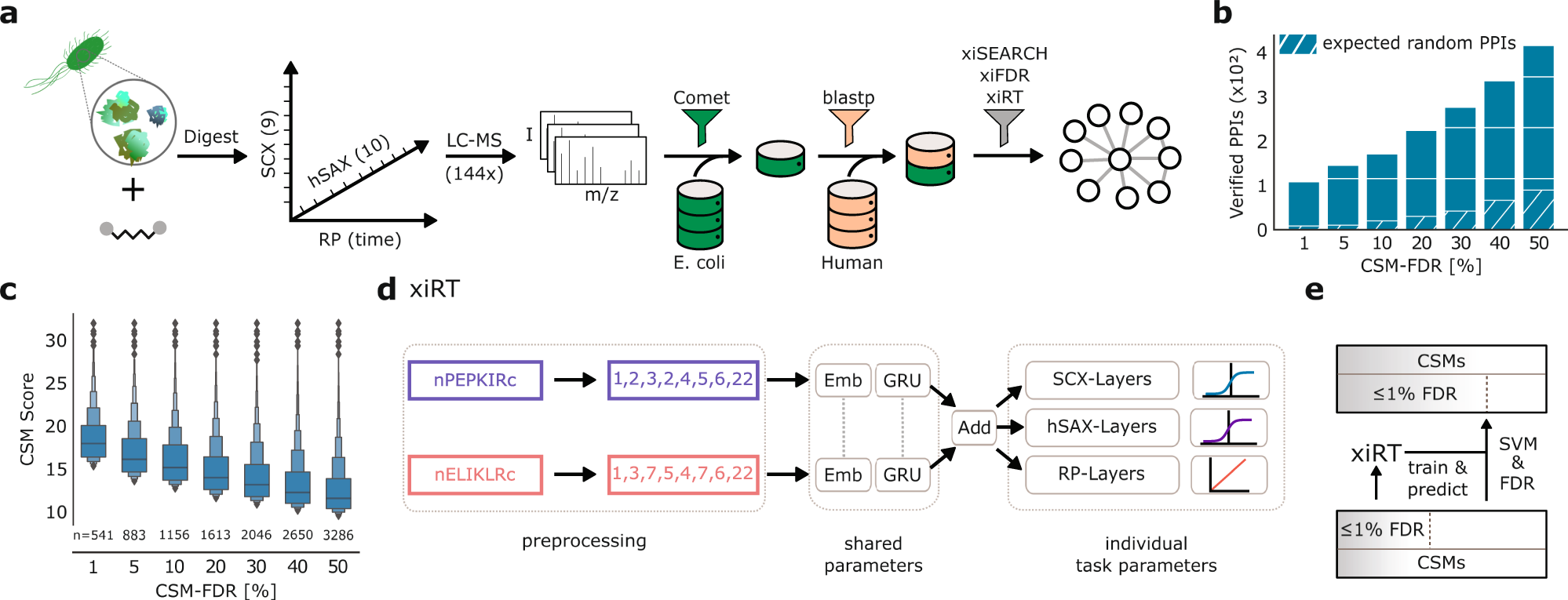
Retention time prediction using neural networks increases identifications in crosslinking mass spectrometry | Nature Communications